AI Innovations in Molecular Structure Prediction Beyond Proteins
Written on
Chapter 1: Expanding the Horizons of Molecular AI
Recent advancements in frontier molecular AI models such as RFAA, the next iteration of AlphaFold, and Umol signal a transformative shift in predicting molecular structures beyond just proteins. Although hurdles like processing speed and reliance on predicted structures remain, these innovations promise to significantly enhance drug development and computational drug design. Keep reading to learn more about these exciting developments.
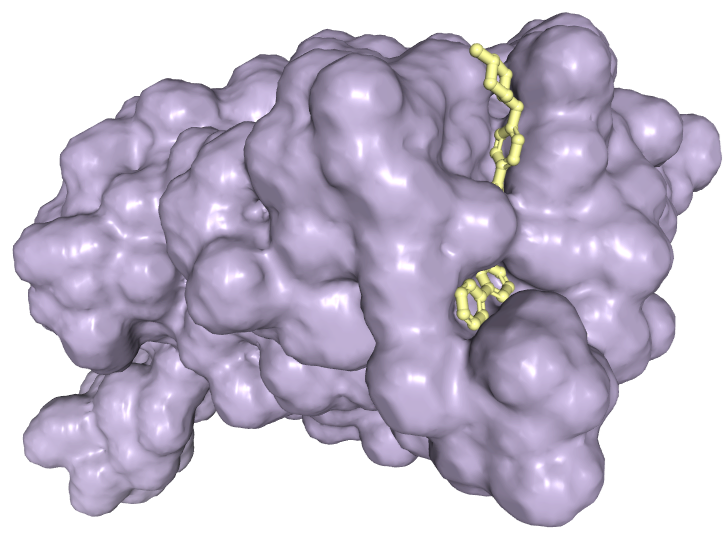
Understanding the complex relationships between proteins and small molecules is vital for progress in both basic biology and drug discovery. Traditional experimental methods, while dependable, tend to be time-consuming and costly. On the other hand, computational approaches are more economical but often struggle with accuracy. This has led to a growing interest in AI-driven solutions, initially focused on modeling 3D structures of proteins. However, proteins represent only a small portion of the vast array of molecular entities that constitute biological systems.
In this article, I will briefly discuss three state-of-the-art models derived from AlphaFold 2 and other contemporary AI tools for protein structure prediction. These models push the boundaries of molecular structure prediction, enabling the prediction of not just proteins but also their interactions with various biologically relevant molecules. The groundbreaking models—RoseTTAFold All-Atom (RFAA), the next generation of AlphaFold, and Umol—demonstrate the potential for AI to redefine the landscape of computational drug design and discovery.
Section 1.1: The Urgency for Faster Drug Development
Creating new pharmaceuticals is not only intellectually demanding but also requires significant financial and time investments. Any acceleration or automation of the drug development process can help overcome these challenges. A critical component is understanding the intricate chemical and physical interactions between proteins and small molecules. Traditionally, this has been achieved through experimental structures or predictive modeling of three-dimensional structures, both of which have limitations. AI-driven solutions could revolutionize this process, akin to the impact AlphaFold 2 had on conventional protein structure prediction.
In a notable competition to launch the first "AI model for all biology," three groups—DeepMind, the Baker lab, and a collaboration between Microsoft and the Noe lab—each introduced pioneering models within just one month. Let’s delve into these innovations.
Subsection 1.1.1: RoseTTAFold All-Atom (RFAA)
RFAA, developed by the Institute of Protein Design (primarily D. Baker’s lab), is capable of managing biological assemblies that include proteins, DNA, RNA, small molecules, and metal ions. This model incorporates diffusion, enabling it to design proteins that interact with various molecules. RFAA utilizes a combination of sequence-based representations for proteins and nucleic acids along with atomic graph structures for small molecules and amino acid modifications, providing a robust solution for protein-small molecule docking challenges.
Section 1.2: Next Generation AlphaFold
Building upon the success of AlphaFold 2, DeepMind is advancing the next generation of AlphaFold, which aims to predict protein structures in conjunction with small molecules. While specific details remain under wraps, the results showcased on their website suggest a potential to replace or enhance traditional methods for predicting protein-small molecule interactions. Similar to RFAA, this new version of AlphaFold will enable simultaneous predictions of protein structures and the binding poses of small molecules.
Chapter 2: Umol - A New Era in Predictive Modeling
Umol, created by the Bryant and Noe labs (the latter supported by Microsoft), allows for the prediction of fully flexible, all-atom structures of protein-ligand complexes directly from a sequence alignment of the protein and a SMILES representation of the ligand. Although it operates slower than traditional docking methods, Umol exceeds RoseTTAFold-AA and classical docking techniques in specific accuracy metrics. Notably, Umol is readily accessible via a Google Colab notebook for immediate use.
Conclusion: A Bright Future for AI in Drug Discovery
The new AI models facilitate an unbiased examination of molecular flexibility, contrasting with traditional docking programs that require initial X-ray structures and predefined ligand conformations. Nevertheless, these modern AI frameworks currently lack the ability to incorporate experimental structures, and their slower speeds can hinder large-scale virtual screening applications. However, given their recent introduction, there is considerable room for enhancement. The intense competition among developers indicates a promising future, with AI-driven drug discovery inching closer to becoming a reality.
References
For a more in-depth exploration, including links to original preprints and related blog entries, visit:
- AlphaFold and Similar AI Models Go All Atoms, Paving New Roads to Drug Development
- Understanding interactions between proteins and small molecules is crucial for advancing fundamental biology and drug...
nexco.ch
www.lucianoabriata.com - I write about a variety of topics, including nature, science, technology, and programming. Subscribe for my latest stories via email. For consultations on small projects, check my services page. You can contact me here or leave a tip here.